As an official AI partner of the Central TB Division (CTD), we are developing multiple interventions across the TB care cascade and helping India’s National TB Elimination Programme (NTEP) become AI-ready.

Line Probe Assay tests diagnose Drug-resistant TB cases (DR-TB). Errors in interpretation, manual entry of results in LPA tests, and a prolonged turnaround time cause delays in initiating TB treatment. We use AI to interpret the results of the LPA test to determine drug resistance to TB. Each LPA strip encodes the drug resistance pattern of the patient via a series of activated (dark) and inactivated (light) bands corresponding to different regions of the genome of the Tuberculosis bacterium. The AI problem consists of identifying this band pattern and then employing a set of rules to determine the specific type of drug resistance.
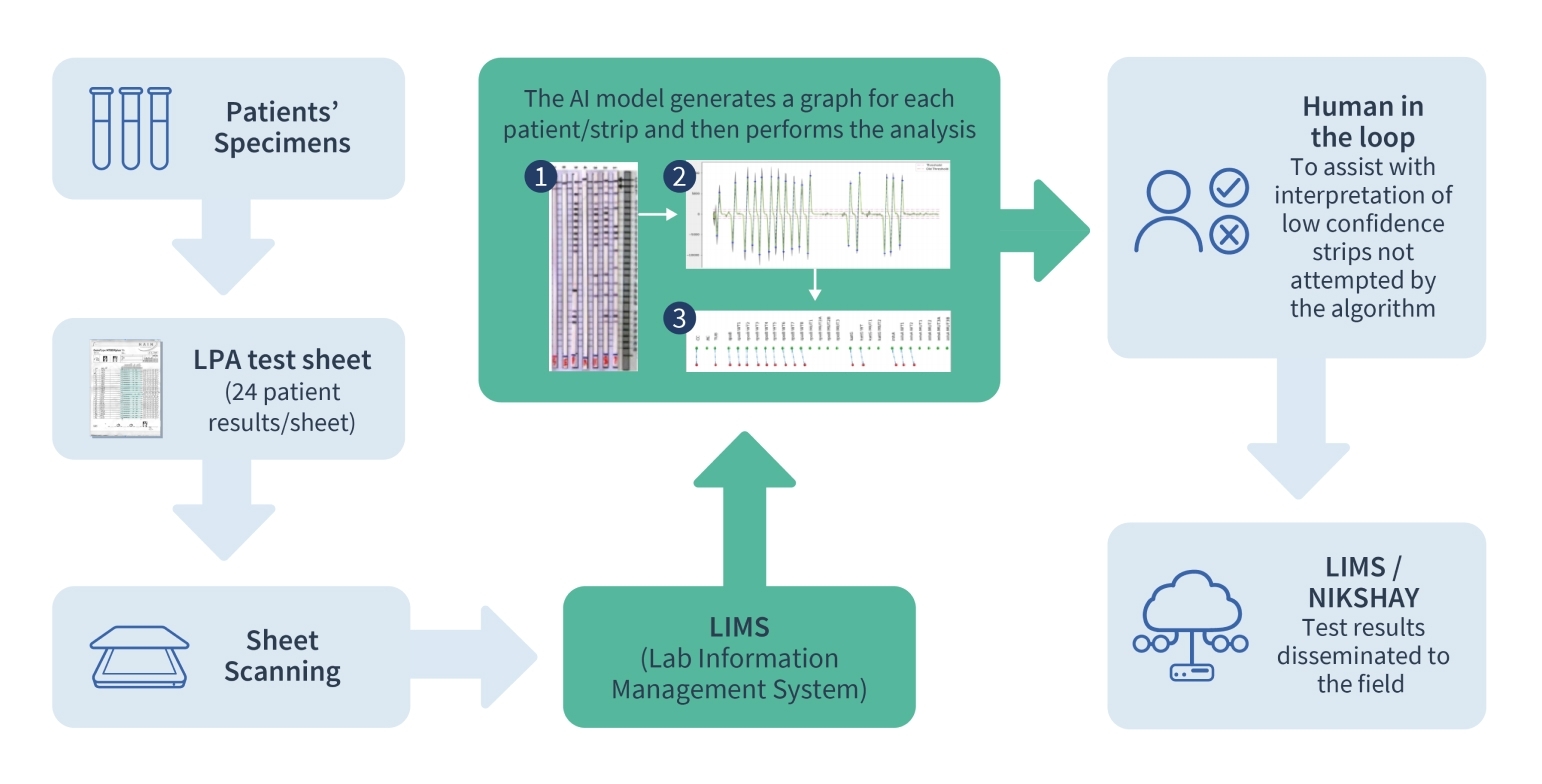
The challenging aspects of this problem lie in visually isolating each strip on a (possibly crumpled) piece of paper and then identifying the bands and their sequence. This is carried out using both classical computer vision techniques that detect activated bands via edge detection and match the band pattern against reference templates. We are working to develop end-to-end supervised deep learning techniques to address this multi-task problem, with a human-in-the-loop as an integral part of the AI ecosystem, along with novel data augmentation techniques.
Treatment success for TB patients depends on their adherence to the treatment regimen. An extreme form of non-adherence is lost to follow-up (LFU), which is defined as a TB patient whose treatment was interrupted for one or more consecutive months (s). This is a risk prediction project – using a set of patient indicators like age, gender, location, the time interval between diagnosis and treatment initiation, etc., collected at treatment initiation time from TB patients, we carry out advance prediction of whether or not the patient will eventually complete treatment.
The AI employs an ensemble of models trained on data corresponding to treatment outcomes for TB patients. The data is feature-engineered – a vast number of categorical indicators are encoded in different ways. New features, for example, those that represent patient migratory behavior, are created by applying specific data transformations. These models were rigorously evaluated on a dataset of patients as part of a blind, prospective evaluation process.
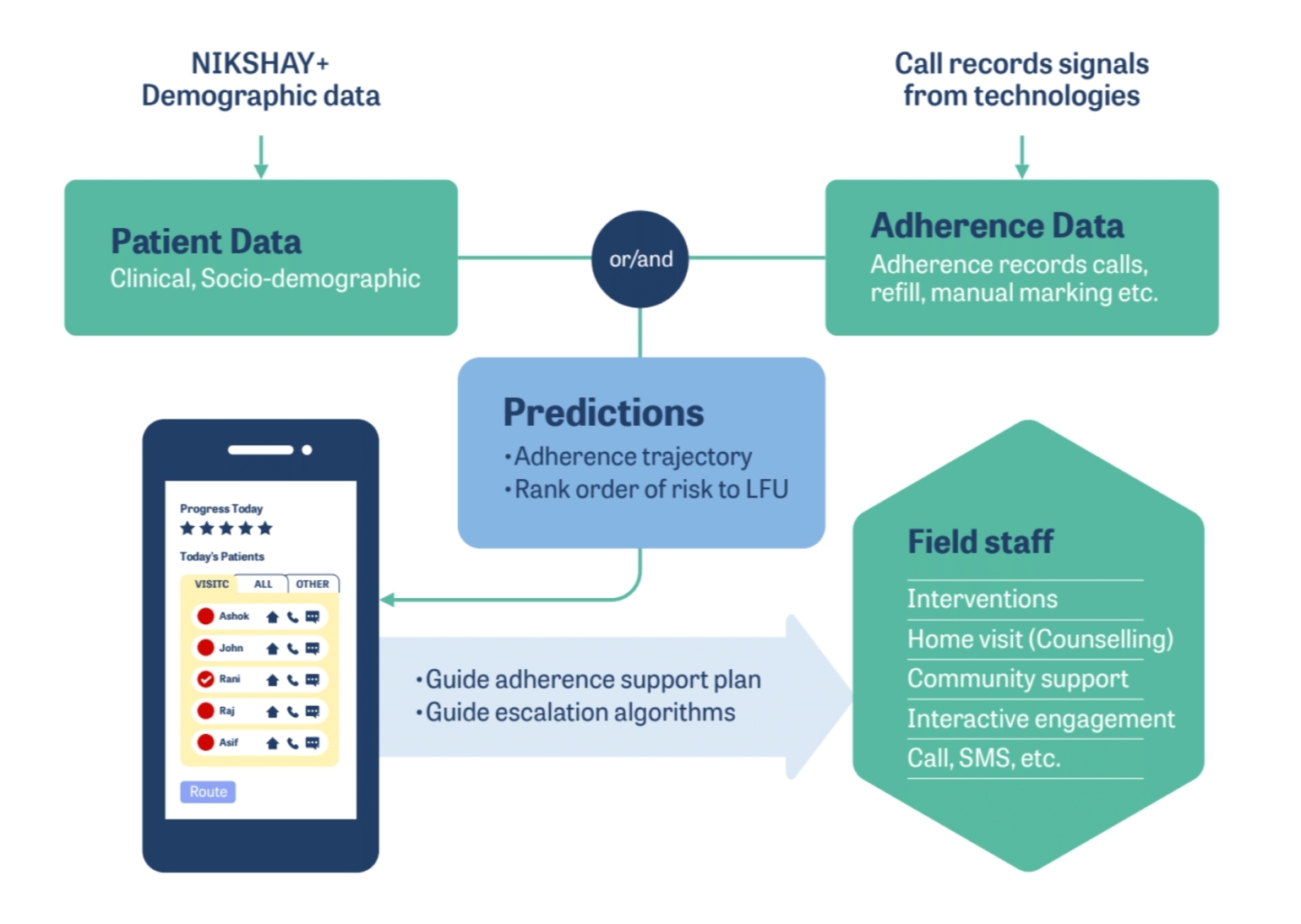
The principal AI challenge here lies in the early prediction of treatment dropoff. Several factors govern treatment dropoff, many of which are dynamic; therefore, early prediction of dropoff is a complex problem. Our AI solution, however, yields accuracies that are more than twice as high as those of the best rule-based decision-making systems and fair across many important cohorts like gender, public vs. private treatment facilities, and month of treatment initiation. We are also working on interpretability methods to ensure that predictions are explainable in terms of the underlying indicators.
A large number of TB patients go unreported every year, and one major reason for this is the limited accessibility and availability of an inexpensive screening tool that is easy to use at the point of care. The screening tools that are currently used are expensive and require special skills and laboratory setups. At a global level, a major priority for TB diagnostic research is to have a rapid, non-invasive, and easy-to-use point-of-care tool for the screening of TB.
Cough is one of the predominant presenting symptoms in cases of pulmonary tuberculosis. Specific characteristics in the cough sound signatures of TB patients are likely to correspond with TB indicators, and screening patients on this basis may help to guide high-probability cases toward early diagnosis and healthcare interventions.
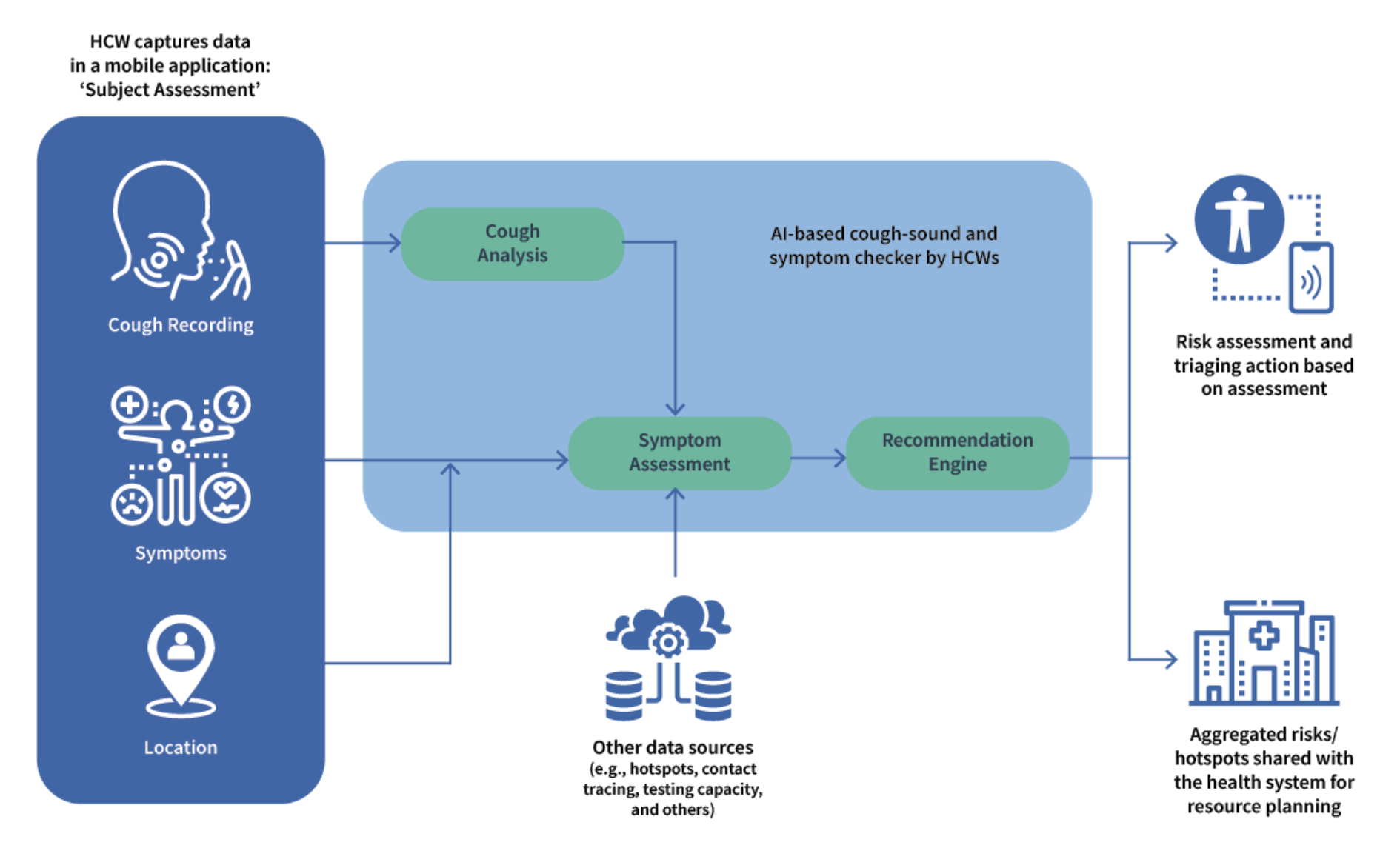
We have developed two AI-powered apps as part of our Cough Against TB solution: one used by healthcare workers to screen TB cases in health facilities and community settings and the other to enable self-screening by individuals in the general population in the comfort of their homes. Our AI model is being trained on data that includes cough and voice sounds and symptoms collected from TB-positive and TB-negative cases. The audio inputs are converted to spectrograms, which, along with symptomatic data, are used as inputs for a deep neural network that learns to predict the likelihood of an individual being a pulmonary TB patient.
A few risk factors and indicators that can influence TB incidence include undernourishment, diabetes, HIV infection, alcohol use disorders, and smoking. Reasons for mortality during treatment of TB have been reported to be due either to extensive TB and complications that come with it or to certain comorbidities. Our AI solution leverages the differentiated care model to mitigate mortality among TB patients who access healthcare facilities in the public sector. Our data collection application, TB-Triage, will manage and support risk prediction, patient stratification, and intervention data capturing.
An AI model is being developed to assess 16 variables (BMI, MUAC, pulse rate, temperature, blood pressure, respiratory rate, oxygen saturation, hemoglobin, icterus, edema, HIV status, RBC, TWC, chest X-ray, hemoptysis, general conditions) and arrive at a machine-driven scoring methodology to enhance the existing system.
The potential users of this app and our solutions are clinical health officers and medical officers who may screen patients, conduct basic clinical assessments, carry out a risk-based stratification of patients, and provide appropriate care to patients identified as high-risk.
Chest X-ray is the leading screening tool for pulmonary TB. However, a significant number of patients still do not undergo screening. This highlights the need for a portable, point-of-care screening system with automated readout. Our USG solution aims to demonstrate that abnormal features found in chest ultrasound scans can be used as signals to diagnose TB using AI. Building upon previous works, we first show that the chest ultrasound scans of TB patients indeed contain distinctive features that are discernible to the radiologist’s eye. We formulate the AI task to detect these abnormal features in an automated fashion and predict the likelihood of the patient being TB-positive. Through deep learning, we identify abnormal features within individual frames of a USG video scan, followed by frame-level aggregation to make a video or patient-level prediction for TB.
A challenging aspect of this solution is ensuring that data collection for building the AI model happens unbiasedly, i.e., USG scans are collected in the same protocol regardless of whether the patient has TB or not. We accomplish this by recommending ‘lawn-mower’ style complete-chest scans for all patients and localized scans that are carried out in equal proportion for TB and non-TB subjects.

© 2025 Wadhwani AI
ROLES AND RESPONSIBILITIES
An ML Engineer at Wadhwani AI will be responsible for building robust machine learning solutions to problems of societal importance; usually under the guidance of senior ML scientists, and in collaboration with dedicated software engineers. To our partners, a Wadhwani AI solution is generally a decision making tool that requires some piece of data to engage. It will be your responsibility to ensure that the information provided using that piece of data is sound. This not only requires robust learned models, but pipelines over which those models can be built, tweaked, tested, and monitored. The following subsections provide details from the perspective of solution design:
Early stage of proof of concept (PoC)
Late PoC
This is early to mid-stage of AI product development
Post PoC
Responsibilities during production deployment
We realize this list is broad and extensive. While the ideal candidate has some exposure to each of these topics, we also envision great candidates being experts at some subset. If either of those cases happens to be you, please apply.
DESIRED QUALIFICATIONS
Master’s degree or above in a STEM field. Several years of experience getting their hands dirty applying their craft.
Programming
ROLES AND RESPONSIBILITIES
As an ML Scientist at Wadhwani AI, you will be responsible for building robust machine learning solutions to problems of societal importance, usually under the guidance of senior ML scientists. You will participate in translating a problem in the social sector to a well-defined AI problem, in the development and execution of algorithms and solutions to the problem, in the successful and scaled deployment of the AI solution, and in defining appropriate metrics to evaluate the effectiveness of the deployed solution.
In order to apply machine learning for social good, you will need to understand user challenges and their context, curate and transform data, train and validate models, run simulations, and broadly derive insights from data. In doing so, you will work in cross-functional teams spanning ML modeling, engineering, product, and domain experts. You will also interface with social sector organizations as appropriate.
REQUIREMENTS
Associate ML scientists will have a strong academic background in a quantitative field (see below) at the Bachelor’s or Master’s level, with project experience in applied machine learning. They will possess demonstrable skills in coding, data mining and analysis, and building and implementing ML or statistical models. Where needed, they will have to learn and adapt to the requirements imposed by real-life, scaled deployments.
Candidates should have excellent communication skills and a willingness to adapt to the challenges of doing applied work for social good.
DESIRED QUALIFICATIONS